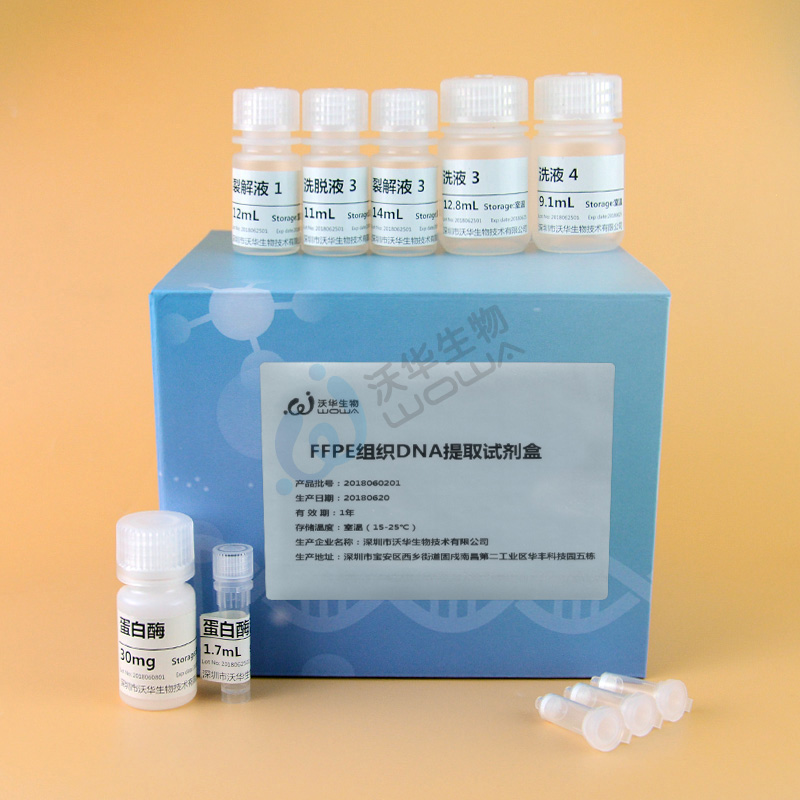



Product Name
Common name: Blood DNA KitFFPE Tissue DNA extraction Kit (column extraction type)
Trade name: Blood DNA KitFFPE Tissue DNA extraction Kit (column extraction type)
Model: ZT-006
Blood DNA KitFFPE Tissue DNA extraction Kit (column extraction type)
User Manual
Product Name
Common name: Blood DNA KitFFPE Tissue DNA extraction Kit (column extraction type)
Trade name: Blood DNA KitFFPE Tissue DNA extraction Kit (column extraction type)
Model: ZT-006
Intended use
This kit uses a centrifugal adsorption column that can specifically bind nucleic acids and a unique buffer system to extract nucleic acids from tissue samples. The silicon matrix material used in the centrifugal adsorption column can efficiently and specifically adsorb nucleic acids, and combined with a unique buffer system, it can maximally remove impurities such as proteins and other organic compounds in cells. The extracted nucleic acid fragments are complete, highly purified, and of stable and reliable quality. The nucleic acid purified using this kit is suitable for various conventional experiments such as enzyme digestion, PCR, library construction, Southern hybridization, and experiments that require high-quality nucleic acid such as chip hybridization and high-throughput sequencing. This product is expected to be used by professionals, such as technicians and doctors trained in molecular biology technology.
Inspection principle
DNA (including genomic DNA and Mitochondrial DNA) can be directly extracted from tissues, and samples can be fixed in Formaldehyde#Forms or embedded in paraffin. The unique lysis buffer system, purified DNA with high concentration, high purity, and good integrity, meets the experimental requirements of chip hybridization and high-throughput sequencing. Adopting the principle of silica gel membrane adsorption, there is no need for phenol chloroform.
【 Main components 】
|
Product composition |
Specifications (50 people) |
|
Cracking liquid 3 |
14mL × one |
|
Cracking liquid 1 |
12mL × one |
|
Adsorption column 1 |
50 |
|
Lotion 3 |
12.8 mL × one |
|
Lotion 4 |
9.1 mL × one |
|
Eluent 3 |
11mL × one |
|
Protease solution |
1.7mL × one |
|
protease |
1 bottle |
Customers are required to prepare their own items, including: 96-100% ethanol, RNase A (100 mg/ml), PBS, xylene, gun head with filter element, metal bath that can be heated to 58 ° C/90 ° C/70 ° C and can be mixed evenly, centrifuge (capable of placing 1.5ml and 2ml centrifuge tubes), vortex oscillator.
Storage conditions and validity period
The reagent kit can be stored for 12 months under dry conditions at room temperature (15-25 ℃). If the room temperature exceeds 25 ℃, adsorption column 1 and protease can be stored at 2-8 ℃.
Reagent preparation
Before use, add 96-100% ethanol to Lotion 3 and Lotion 4, and add 17ml 96-100% ethanol to Lotion 3 bottles; Add 21ml of 96-100% ethanol to 4 bottles of washing solution. Add 1.5ml of protease solution to the protease bottle for dissolution (store the dissolved protease at 2-8 ℃).
Sample requirements
Extraction of FFPE tissue DNA (including genomic DNA and Mitochondrial DNA)
The sample shall be fixed in Formaldehyde#Forms and embedded in paraffin. To limit the degree of DNA breakage, it is necessary to ensure that:
1. Fix tissue samples with 4-10% Formaldehyde#Forms as soon as possible after surgical resection
2. The fixed time used is 14-24 hours (longer fixed time leads to more severe DNA breakage, resulting in poor performance in downstream analysis).
3. Thoroughly dehydrate the sample before embedding (residual Formaldehyde#Forms can inhibit the digestion of Proteinase K).
4. The raw material used for DNA purification should be fresh FFPE tissue slices, and the number of slices in one preparation should not exceed 8, and the thickness of each slice should not exceed 10 μ Three conditions: m and a surface area not exceeding 250 mm2. If you do not have relevant information on the properties of the raw materials, we suggest starting each preparation with no more than 3 slices. Depending on DNA yield and purity, up to 8 slices may be used in subsequent preparations.
Pre treatment of paraffin embedded tissue samples:
1. Use the Scalpel to trim the excess paraffin in the sample block.
2. Cut no more than 4 slices with a thickness of 10 μ M; Or cut no more than 8 slices with a thickness of 5 μ M. If the surface of the sample has been exposed to air, discard the first 2-3 slices. Immediately place each slice in a 1.5 or 2 ml microcentrifuge tube (not provided).
3. Add 1 ml of xylene to the sample. Close the lid and swirl vigorously for 10 seconds. Centrifuge at full speed for 2 minutes at room temperature (15-25 ℃). Remove the supernatant. No sediment can be removed.
4. Add 1 ml of ethanol (96~100%) to the precipitate and vortex mix well. Ethanol extraction of residual xylene in the sample. Centrifuge at full speed for 2 minutes at room temperature (15-25 ℃). Remove the supernatant. No sediment can be removed. Carefully remove any residual ethanol using a thin Pipette tip.
Note: If necessary, this step can be repeated once.
5. Open the tube and incubate at a temperature not exceeding 37 ℃ for 10 minutes or until all residual ethanol evaporates dry.
6. Add 180 μ Resuspended precipitation in cracking solution 1. Join 25 μ L protease, vortex mix well.
7. Incubate and digest at 58 ℃ for 1-3 hours or until the sample is fully digested (in most cases, it can be completely dissolved within 1 hour, and sometimes overnight may be necessary). It is recommended to place the sample on a shaking table or other instruments during digestion and keep it continuously mixed. Then extract the nucleic acid according to the following steps for nucleic acid extraction.
Pretreatment of Formaldehyde#Forms fixed tissue samples:
1. Cut no more than 25mg of tissue or 10mg of spleen tissue and wash the sample twice with PBS to remove the fixative.
2. Choose one of the following three methods to process the sample (b, c): a. Cut the tissue into small pieces and add 220 μ Cracking liquid 1; b. Place the tissue in liquid nitrogen and thoroughly grind with a mortar and pestle. Then pour the tissue powder and liquid nitrogen into a 1.5 milliliter microcentrifuge tube. Allow liquid nitrogen to evaporate (but do not allow tissue to thaw) and add 220 μ Cracking liquid 1; c. Organize and 80 μ Add PBS to the homogenizer for homogenization, and after tissue homogenization, add 140 μ Cracking liquid 1.
3. Add 25 μ L protease, vortex mix well, incubate and digest at 58 ℃ for 1-3 hours or until the sample is fully digested. It is recommended to place the sample on a shaking table or other instruments during digestion and keep it continuously mixed.
4. Then extract the nucleic acid according to the following steps for nucleic acid extraction.
【 Precautions 】 (Please be sure to read this notice before using this reagent kit):
1. The sample should avoid repeated freeze-thaw, otherwise it may result in smaller extracted DNA fragments and a decrease in extraction volume.
If there are precipitates in cracking solution 1 and 3, they can be dissolved again in a 70 ℃ water bath, shaken well, and then used.
3. RNA can inhibit some downstream enzyme reactions, but it will not inhibit PCR. To remove RNA residues, you need to prepare your own RNase A (100 mg/ml) solution.
4. Balance the sample and all buffer solutions to room temperature.
[Operation Steps]
1. Instantaneous centrifugation, adding 4 μ Mix RNase A (100 mg/ml) and take a warm bath at room temperature (15-25 ℃) for 2 minutes; If RNA removal is not necessary, skip to the next step without doing so.
Incubate and digest at 2.90 ℃ for 1 hour, centrifuge instantly, and add 200 μ L cracking solution 3, thoroughly mix well.
3. Instantaneous centrifugation, adding 200 μ Ethanol (96~100%), cover with lid, vortex oscillate and mix for 15 seconds.
Note: If the room temperature exceeds 25 ° C, cool the ethanol on ice before adding it to the tube.
4. Instantaneous centrifugation, add to the adsorption column, centrifuge at a speed of over 6000g for 1 minute, place the adsorption column in a clean 2 ml collection tube, and discard the collection tube containing the effluent.
Note: 6000g centrifugation will only reduce noise, and centrifugation at a higher speed will not affect DNA extraction.
5. Add 500 μ Centrifuge at a rate of 6000g of washing solution 3 (with ethanol added) for 1 minute, place the adsorption column in a clean 2 ml collection tube, and discard the collection tube containing the effluent.
6. Add 500 μ Centrifuge solution 4 (with ethanol added) at a rate of 6000g for 1 minute, place the adsorption column in a clean 2 ml collection tube, and discard the collection tube containing the effluent.
Centrifuge at a speed of 7.2000g for 3 minutes.
8. Replace the centrifuge tube and add 50 drops to the middle position of the adsorption membrane μ L -200 μ Elute 3, warm bath at room temperature for 5 minutes, centrifuge at a rate of 20000gMinutes.
